Propensity Score Diagnostics
Lucy D’Agostino McGowan
Wake Forest University
2020-07-29 (updated: 2020-07-29)
1 / 35
Checking balance
- Love plots (Standardized Mean Difference)
- ECDF plots
2 / 35
Standardized Mean Difference (SMD)
d=¯xtreatment−¯xcontrol√s2treatment+s2control2
3 / 35
SMD in R
1
Create a "design object" to incorporate the weights
library(survey)svy_des <- svydesign( ids = ~ 1, data = df, weights = ~ wts)4 / 35
SMD in R
2
Calculate the unweighted standardized mean differences
library(tableone)library(tidyverse)smd_table_unweighted <- CreateTableOne( vars = c("confounder_1", "confounder_1", ...), strata = "exposure", data = df, test = FALSE)5 / 35
SMD in R
3
Calculate the weighted standardized mean differences
smd_table <- svyCreateTableOne( vars = c("confounder_1", "confounder_1", ...), strata = "exposure", data = svy_des, test = FALSE)6 / 35
SMD in R
3
Calculate the weighted standardized mean differences
smd_table <- svyCreateTableOne( vars = c("confounder_1", "confounder_1", ...), strata = "exposure", data = svy_des, test = FALSE)7 / 35
SMD in R
4
Stick these together in a data frame
plot_df <- data.frame( var = rownames(ExtractSmd(smd_table)), Unadjusted = as.numeric(ExtractSmd(smd_table_unweighted)), Weighted = as.numeric(ExtractSmd(smd_table))) %>% pivot_longer(-var, names_to = "Method", values_to = "SMD")8 / 35
SMD in R
4
Stick these together in a data frame
plot_df <- data.frame( var = rownames(ExtractSmd(smd_table)), Unadjusted = as.numeric(ExtractSmd(smd_table_unweighted)), Weighted = as.numeric(ExtractSmd(smd_table))) %>% pivot_longer(-var, names_to = "Method", values_to = "SMD")rownames(EXtractSMD(smd_table))#> [1] "confounder_1" "confounder_2"9 / 35
SMD in R
4
Stick these together in a data frame
plot_df <- data.frame( var = rownames(ExtractSmd(smd_table)), Unadjusted = as.numeric(ExtractSmd(smd_table_unweighted)), Weighted = as.numeric(ExtractSmd(smd_table))) %>% pivot_longer(-var, names_to = "Method", values_to = "SMD")as.numeric(ExtractSmd(smd_table_unweighted))#> [1] 0.160 0.17710 / 35
SMD in R
4
Stick these together in a data frame
plot_df <- data.frame( var = rownames(ExtractSmd(smd_table)), Unadjusted = as.numeric(ExtractSmd(smd_table_unweighted)), Weighted = as.numeric(ExtractSmd(smd_table))) %>% pivot_longer(-var, names_to = "Method", values_to = "SMD")as.numeric(ExtractSmd(smd_table))#> [1] 0.002 0.00711 / 35
SMD in R
4
Stick these together in a data frame
plot_df <- data.frame( var = rownames(ExtractSmd(smd_table)), Unadjusted = as.numeric(ExtractSmd(smd_table_unweighted)), Weighted = as.numeric(ExtractSmd(smd_table))) %>% pivot_longer(-var, names_to = "Method", values_to = "SMD")12 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()13 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()14 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()15 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()16 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()17 / 35
SMD in R
5
Plot them! (in a Love plot!)
ggplot(data = plot_df, mapping = aes(x = var, y = SMD, group = Method, color = Method)) + geom_line() + geom_point() + geom_hline(yintercept = 0.1, color = "black", size = 0.1) + coord_flip()18 / 35
Love plot
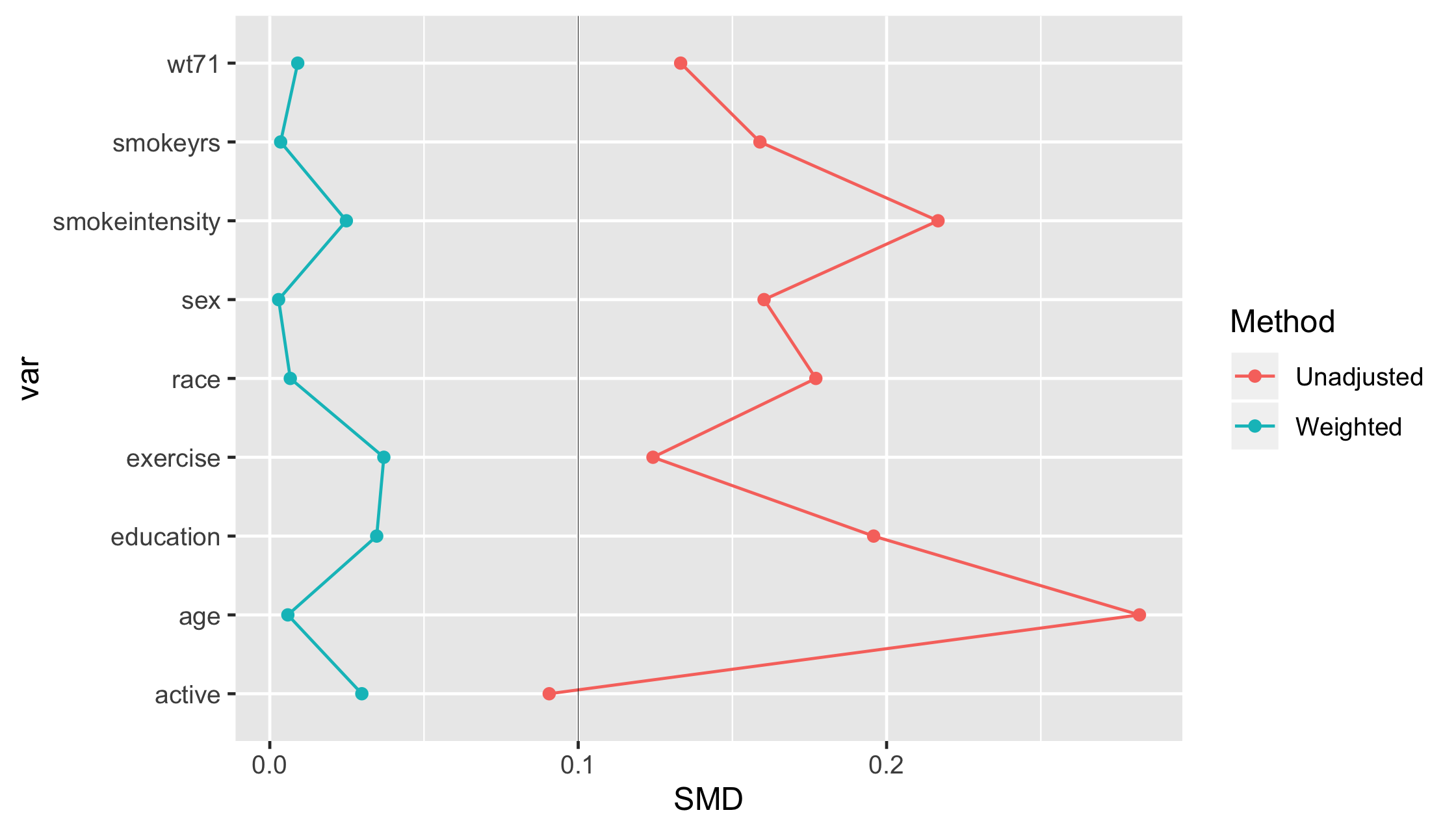
19 / 35
Your turn 1
07:00
- Create a Love Plot for the propensity score weighting you created in the previous exercise
20 / 35
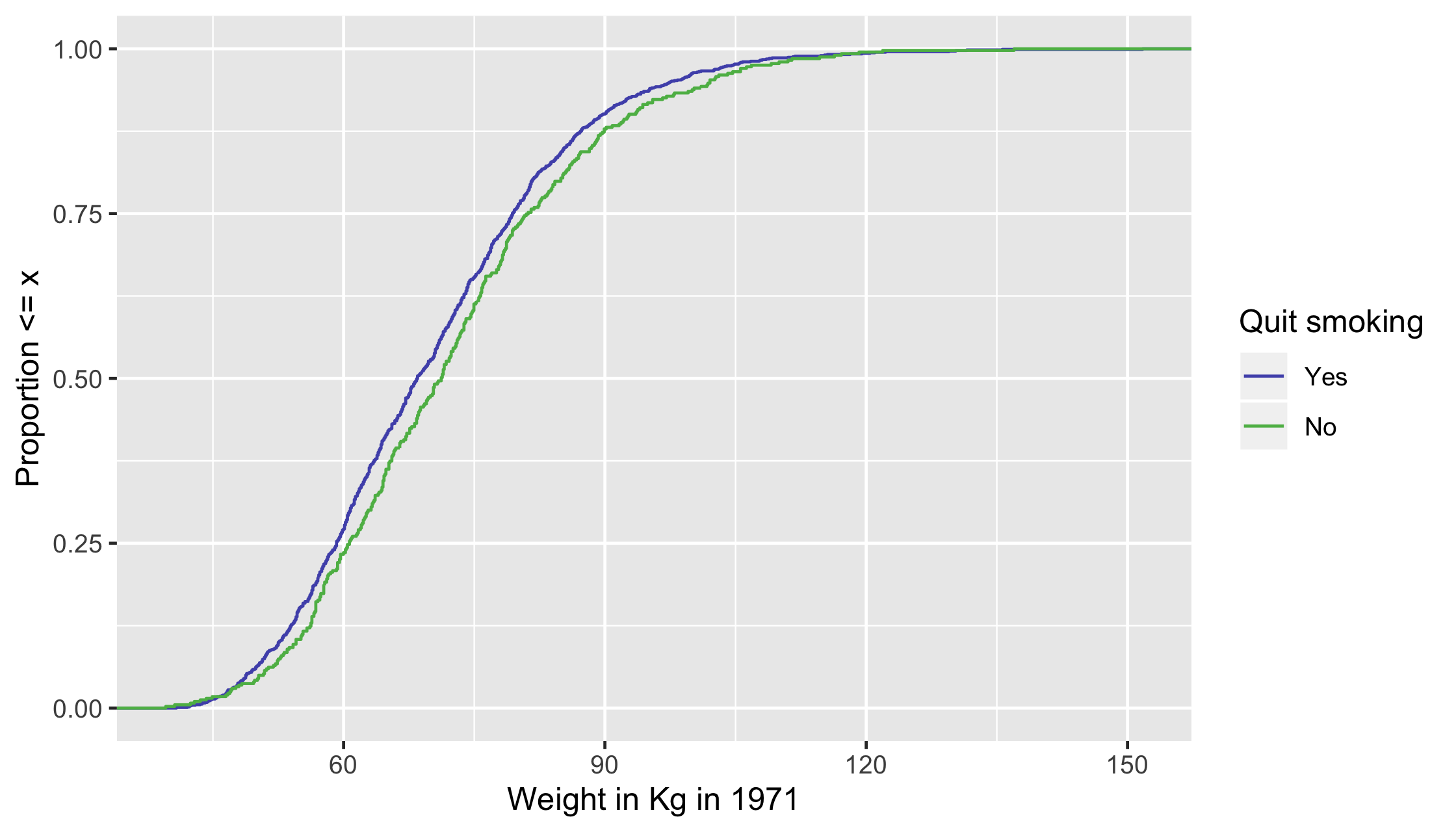
ECDF
For continuous variables, it can be helpful to look at the whole distribution pre and post-weighting rather than a single summary measure
21 / 35
Unweighted ECDF
ggplot(df, aes(x = wt71, group = qsmk, color = factor(qsmk))) + stat_ecdf() + scale_color_manual("Quit smoking", values = c("#5154B8", "#5DB854"), labels = c("Yes", "No")) + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")22 / 35
Unweighted ECDF
ggplot(df, aes(x = wt71, group = qsmk, color = factor(qsmk))) + stat_ecdf() + scale_color_manual("Quit smoking", values = c("#5154B8", "#5DB854"), labels = c("Yes", "No")) + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")23 / 35
Unweighted ECDF
ggplot(df, aes(x = wt71, group = qsmk, color = factor(qsmk))) + stat_ecdf() + scale_color_manual("Quit smoking", values = c("#5154B8", "#5DB854"), labels = c("Yes", "No")) + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")24 / 35
Unweighted ECDF
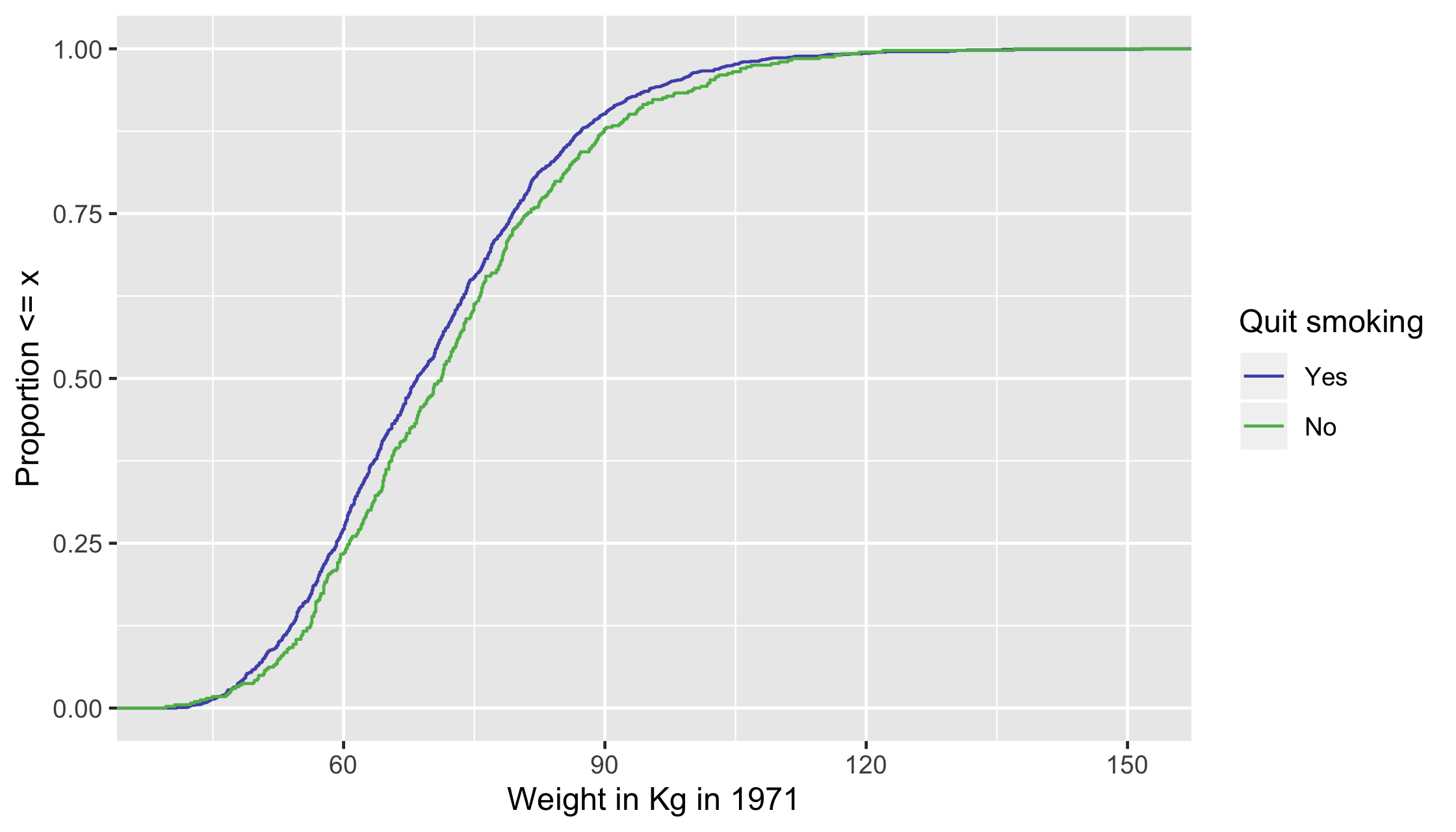
25 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")26 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")27 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")28 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")29 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")30 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")31 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")32 / 35
Weighted ECDF
ecdf_1 <- df %>% filter(qsmk == 1) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ecdf_0 <- df %>% filter(qsmk == 0) %>% arrange(wt71) %>% mutate(cum_pct = cumsum(w_ate) / sum(w_ate))ggplot(ecdf_1, aes(x = wt71, y = cum_pct)) + geom_line( color = "#5DB854") + geom_line(data = ecdf_0, aes(x = wt71, y = cum_pct), color = "#5154B8") + xlab("Weight in Kg in 1971") + ylab("Proportion <= x")33 / 35
Weighted ECDF
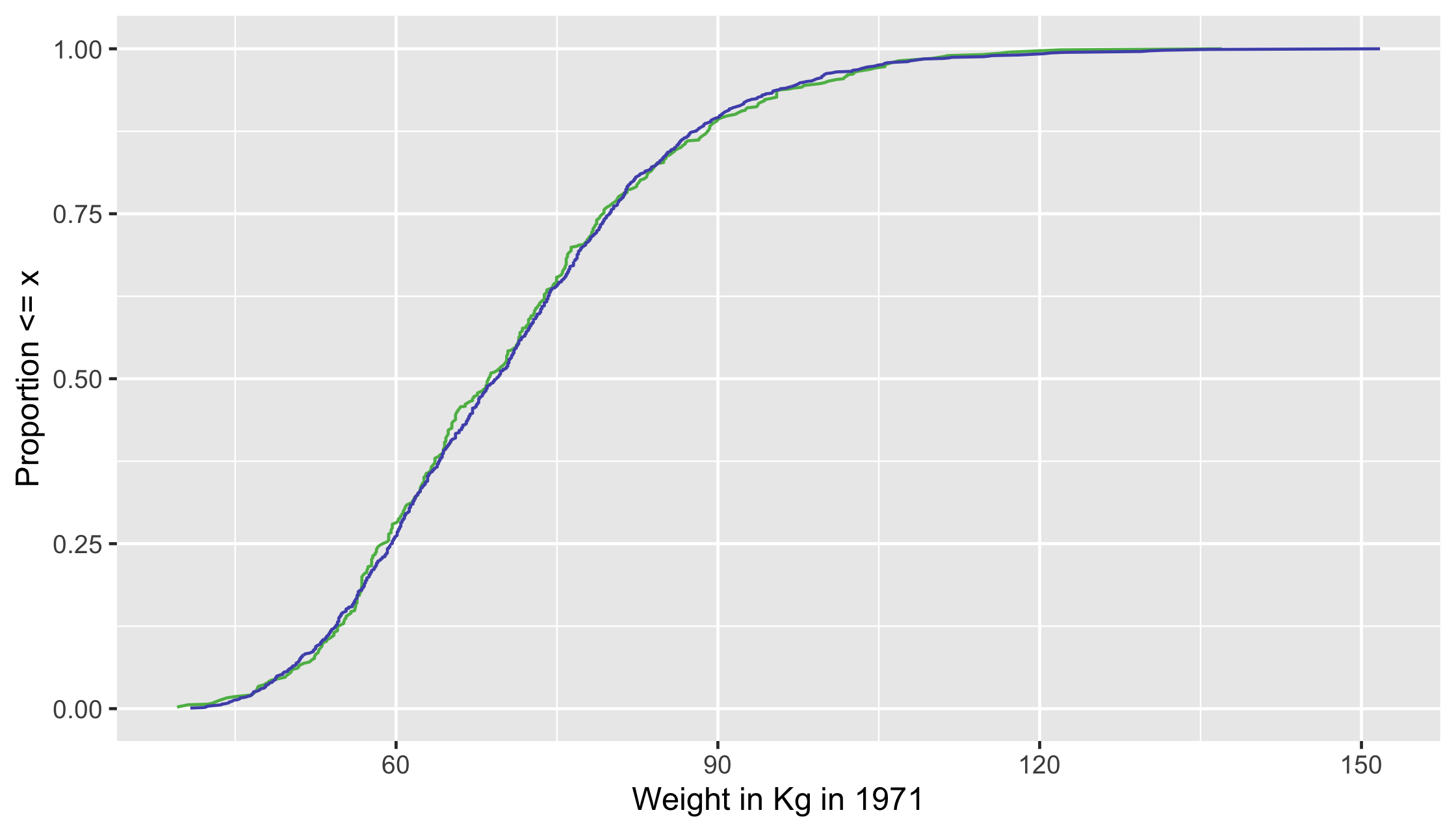
34 / 35
Your turn 2
07:00
- Create an unweighted ECDF examining the
smokeyrsconfounder for those that quit smoking and those that did not - Create a weighted ECDF examining the
smokeyrsconfounder
35 / 35